Abstract
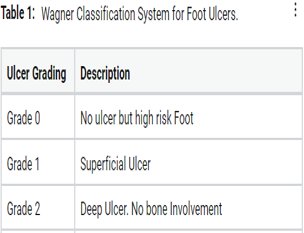
Type II diabetes mellitus is a significant health problem that developed globally. This study was carried out on patients with diabetic foot ulcer (DFU) to assess the bacterial and fungal flora, susceptibility, and drug-resistant isolates and devises an empiric antimicrobial therapy. Clinical data and patient samples were collected from 300 diabetic foot ulcer patients between September 2014, and September 2016 and samples were processed as per CLSI guidelines. Most of the pathogenic isolate recovered according to the Wagner classification system in DFU. The most commonly found isolates in our Study was Pseudomonas aeruginosa (22%), Staphylococcus aureus (15%), Escherichia coli (11%) followed by others. Antimicrobial resistance appears in aerobic, anaerobic as well as candida isolates in our study. Our results show most gram-negative bacteria were sensitive to colistin and tigecycline, and 44% of Gram-negative bacteria were ESBL producers, and among 20% of the gram-negative isolates were Multidrug resistant (MDR) organisms. Proper diagnosis of the causative agents, surveillance monitoring on the susceptibility of the isolates and determining the drugs for the empirical treatment of diabetic foot ulcers will prevent prolonged hospital stay and amputation.
Full text article
Authors

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.