Abstract
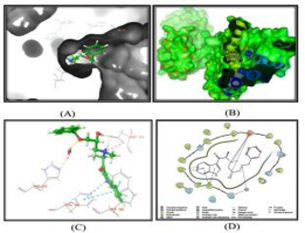
Cancer is a big issue that affects people all over the world. It develops as a result of uncontrolled cell growth. The interaction between developed ligands and thymine phosphorylation was investigated in this study, which was computationally optimized. The aim of this study was to examine the anticancerous activity of designed ligands in thymine phosphorylation (PDB ID: 1UOU) in order to minimize the cost and time required to develop a novel anticancer drug with minimal side effects. All the designed ligands showed mild to excellent binding with proteins. Most of the ligands exhibited better interaction compared to reference compound Tamoxifen with pdb files. Some of the designed ligands among (1-7) in qunoline derivatives and (1-5) in Chalcone derivatives showed excellent docking scores with PDB file (1UOU) of thymine phosphorylation. All the designed ligands and Zinc databases were docked with 1UOU PDB files of protein, and it was found that out of twenty-five designed ligands in Qunoline series, ligand 25 showed the best binding (docking score−8.268) with 1UOU PDB of protein thymine phosphorylation. And that out of ten designed ligands in Chalcone series, ligand K1 showed the best binding (docking score−9.433) with 1UOU PDB of protein thymine phosphorylation. Docked ligand cavity of ligand ku 25 in qunoline series and K 9 in Chalcone series showed important hydrophobic/non-polar residues such as Ile199, Ile316, Trp119, Phe168, Ile198, Cys172, Tyr188, Tyr398, Tyr435, Phe343, Tyr60, Leu328, Leu171, and showed pi-pi interaction with Tyr326. Further wet laboratory studies are continued in our laboratory to confirm and find out the efficiency and activity of target compounds.
Full text article
References
Balakumar, C., Lamba, P., Kishore, D. P., Narayana, B. L., Rao, K. V., Rajwinder, K., Rao, A. R., Shireesha, B., Narsaiah, B. 2010. Synthesis, anti-inflammatory evaluation and docking studies of some new fluorinated fused quinazolines. European journal of medicinal chemistry, 45(11):4904– 4913.
Berry, M., Ahmed, Z., Lorber, B., Douglas, M., Logan, A. 2008. Regeneration of axons in the visual system. Restorative neurology and neuroscience, 26:147–174.
Bhosale, M., Yadav, A., Magdum, C., Mohite, S. 2020. Microwave Assisted Synthesis, Molecular Docking Studies and Anticancer Screening of Some 1, 3, 4-thiadiazole Derivatives. Journal of the University of Shanghai for Science and Technology, 22(11):520.
Bora-Tatar, G., Dayangaç-Erden, D., Demir, A. S., Dalkara, S., Yelekçi, K., Erdem-Yurter, H. 2009. Molecular modifications on carboxylic acid derivatives as potent histone deacetylase inhibitors: Activity and docking studies. Bioorganic and medicinal chemistry, 17(14):5219–5228.
Chou, C. H., Shrestha, S., Yang, C. D., Chang, N. W., Lin, Y. L., Liao, K. W., Huang, W. C., Sun, T. H., Tu, S. J., Lee, W. H., Chiew, M. Y. 2018. miRTarBase update 2018: a resource for experimentally validated microRNA-target interactions. Nucleic acids research, 46(D1):296–302.
Daina, A., Michielin, O., Zoete, V. 2017. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Scientific Reports, 7(1):1–13.
El-Karim, A. S. S., Anwar, M. M., Mohamed, N. A., Nasr, T., Elseginy, A. S. 2015. Design, synthesis, biological evaluation and molecular docking studies of novel benzofuran-pyrazole derivatives as anticancer agents. Bioorganic chemistry, 63:1–12.
Farid, R., Day, T., Friesner, R. A., Pearlstein, R. A. 2006. New insights about HERG blockade were obtained from protein modelling, potential energy mapping, and docking studies. Bioorganic and medicinal chemistry, 14(9):3160–3173.
Feher, M., Williams, C. I. 2010. Reducing docking score variations arising from input differences. Journal of chemical information and modelling, 50:1549–1560.
Gilad, Y., Senderowitz, H. 2014. Docking studies on DNA intercalators. Journal of chemical information and modelling, 54(1):96–107.
Goldman, B. B., Wipke, W. T. 2000. QSD quadratic shape descriptors. 2. Molecular docking using quadratic shape descriptors (QSDock). Proteins: Structure, Function, and Bioinformatics, 38:79–94.
Kim, M. J., Loucks, R. A., Palmer, A. L., Brown, A. C., Solomon, K. M., Marchante, A. N., Whalen, P. J. 2011. The structural and functional connectivity of the amygdala: from normal emotion to pathological anxiety. Behavioural brain research, 223(2):403–410.
Kitchen, D. B. 2004. Docking and scoring in virtual screening for drug discovery: methods and applications. Nature reviews Drug discovery, 3:935–949.
Olivieri, G. L., Sousa, V., Chikhi, L., Radespiel, U. 2008. From genetic diversity and structure to conservation: genetic signature of recent population declines in three mouse lemur species (Microcebus spp.) Biological Conservation, 141(5):1257–1271.
Oshiro, C., Bradley, E. K., Eksterowicz, J., Evensen, E., Lamb, M. L., Lanctot, J. K., Putta, S., Stanton, R., Grootenhuis, P. D. 2004. Performance of 3D-database molecular docking studies into homology models. Journal of medicinal chemistry, 47(3):764–767.
Pang, Y. P., Kozikowski, A. P. 1994. Prediction of the binding sites of huperzine A in acetylcholinesterase by docking studies. Journal of Computer-Aided Molecular Design, 8(6):669–681.
Qin, H. L., Shang, Z. P., Jantan, I., Tan, O. U., Hussain, M. A., Sher, M., Bukhari, S. N. A. 2015. Molecular docking studies and biological evaluation of chalcone based pyrazolines as tyrosinase inhibitors and potential anticancer agents. RSC Advances, 5:46330–46338.
Redecker, C., Leis, M., Leendertse, M., Punie, Y., Gijs- bers, G., Kirschner, P., Stoyanov, S., Hoogveld, B. 2011. The future of learning: Preparing for change. Luxembourg: Publications Office of the European Union.
Ricci, C. G., Netz, P. A. 2009. Docking studies on DNA-ligand interactions: building and application of a protocol to identify the binding mode. Journal of chemical information and modelling, 49(8):1925–1935.
Taylor, S. E. 2010. Mechanisms linking early life stress to adult health outcomes. Proceedings of the National Academy of Sciences, 107:8507–8512.
Vijesh, A. M., Isloor, A. M., Telkar, S., Arulmoli, T., Fun, H. K. 2013. Molecular docking studies of some new imidazole derivatives for antimicrobial properties. Arabian Journal of Chemistry, 6(2):197–204.
Wang, G. 2018. Synthesis, biological evaluation and molecular docking studies of a new series of chalcones containing naphthalene moiety as anticancer agents. Bioorganic chemistry, 76:249–257.
Wang, G., Wei, Y., Qiao, S., Lin, P., Chen, Y. 2018. Generalized inverses: theory and computations. volume 53. Springer.
Welsh, A., Hill, T., Quinlan, H., Robinson, C., May, B. 2008. Genetic assessment of lake sturgeon population structure in the Laurentian Great Lakes. North American Journal of Fisheries Management, 28(2):572–591.
Yongye, A. B., Bender, A., Martínez-Mayorga, K. 2010. Dynamic clustering threshold reduces conformer ensemble size while maintaining a biologically relevant ensemble. Journal of computer-aided molecular design, 24:675–686.
Yuriev, E., Ramsland, P. A. 2013. Latest developments in molecular docking: 2010-2011 in review. Journal of Molecular Recognition, 26:215–239.
Authors

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.